Why Multiply When You're About to Divide?
Problem
The Polymerase Chain Reaction (PCR) is a technique used to amplify the amount of DNA in a sample. During each round of PCR, the DNA double helix is unwound and separated into two separate strands. A new strand is then synthesised against each of these.
How does the DNA content of a sample change with a single round of PCR?
If there is originally 1mg of DNA in the sample, then what mass of DNA will be present after 15 rounds of PCR?
If my sample of 0.5ml with a DNA concentration of 0.1pM, how many rounds of PCR until I have 10$^{12}$ molecules of DNA?
DNA synthesis in PCR is carried out by a Taq DNA polymerase. This can build a new strand of DNA against a template one at a rate of 6000 bases per minute.
If a piece of single stranded DNA has 24 kbases, what is the minimum number of minutes that should be left between rounds of PCR?
If I start with a single 24000 base pair DNA helix, and carry out 10 rounds of PCR, how many bases will have been incorporated into DNA at the end?
What is the point of PCR? Amplification of small quantities of DNA allows it to be analysed using a variety of methods -one of these techniques is restriction mapping. Restriction mapping relies on bacterially-derived restriction enzymes which can cut DNA at specific sequences. Each type of restriction enzyme cuts the DNA at a different but specific sequence. When a DNA helix has been cut using
these enzymes, the lengths of the fragments can be determined by using gel electrophoresis, which separates the molecules on the basis of size. By using different combinations of restriction enzymes to generate different length fragments, a map can be drawn of the original DNA with the location of each type of restriction site.
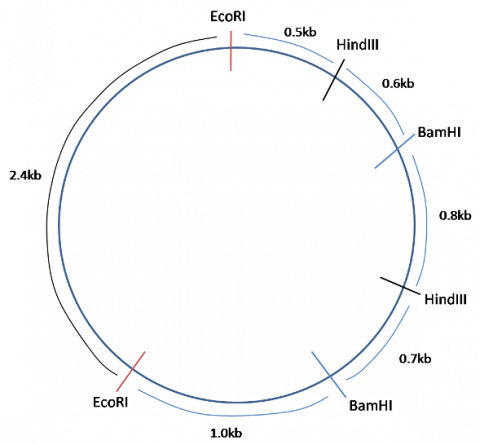
For example, a certain bacterial plasmid (circular DNA molecule) is digested with different combinations of restriction enzymes, and gives the following size fragments:
BamHI: 2.1kb, 1.9kb
HindIII: 3.5kb, 0.5kb
BamHI + HindIII: 1.9kb, 1.3kb, 0.5kb, 0.3kb.
From this data, is it possible to deduce the following map of the plasmid: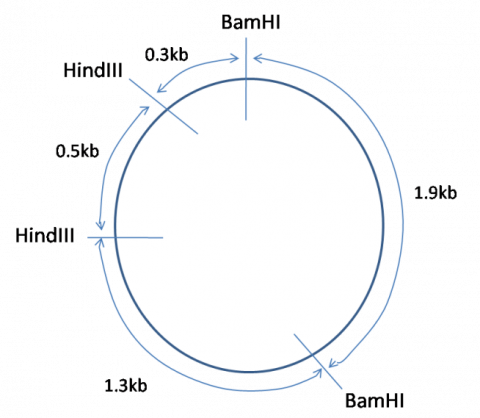
Using the above example as a guide, create a map of the restriction sites on the plasmid from the following data (all fragment lengths in kb):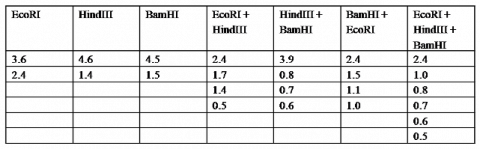
Getting Started
Note that some restriction maps are not unique, in that the data given may allow more than one valid map to be drawn.
Student Solutions
1a) During one round of PCR, the DNA double helix is separated into its constituent strands, and new strands are built up against each of these. The final products are two helices, and so the DNA content of the cell doubles.
b) If there is originally 1mg of DNA in the sample, 30 rounds of PCR, will lead to doubling thirty times:
$Mass_{final} = 0.001 \times 2^{15} = 32.8$g of DNA
c) We must first calculate the number of moles of DNA in the sample:
$Moles = \text{concentration} \times \text{volume} = 0.1 \times 10^{-12} \times 0.5 \times 10^{-3} = 5 \times 10^{-17}$ moles DNA
The number of molecules of DNA is calculated by multiplying moles by Avogadro's number:
$Molecules = 5 \times 10^{-17} \times 6.02 \times 10^{23} = 3.01 \times 10^7$
Thus to work out the number of rounds of PCR necessary:
$3.01 \times 10^7 \times 2^n = 10^{12}$
$2^n = \frac{10^{12}}{3.01 \times 10^7}$
$n = \frac{ln(\frac{10^{12}}{3.01 \times 10^7})}{ln(2)} = 15.02$
Therefore, we require 16 rounds of PCR to exceed $10^{12}$ molecules of DNA.
2a) Number of minutes = $\frac{24000}{6000} = 4$
b) In the first round of PCR, we will incorporate $2 \times 24000 = 48000$ bases into DNA. In the next round there will be four template strands, and so $4 \times 24000 = 96000$ bases will be incorporated into DNA. Thus over a total of ten rounds:
Number of incorporated bases = $ 24000 \times ( 2^1 + 2^2 +2^3 +...+2^{10})$
The part in brackets is a geometric series, which can be summed as follows:
Let $ x = 2^1 + 2^2 +2^3 +...+2^{10}$
Then $2x = 2^2 +2^3 +2^4 +...+2^{11}$
$\therefore 2x -x = 2^{11} - 2$
$\rightarrow x = 2^{11} -2= 2(2^{10} - 1)$
Thus, the number of incorporated bases = $24000 \times 2(2^{10} -1)= 48000(2^{10} -1) = 4.91 \times 10^7$
3) Solving the restriction map is done by partly by trial and error, and partly by building the map up with more and more information until the solution is found: